線形回帰は全ての基本です。
変分推論のロジックがわかったら、エグい計算はライブラリを使って避けたいです。
model
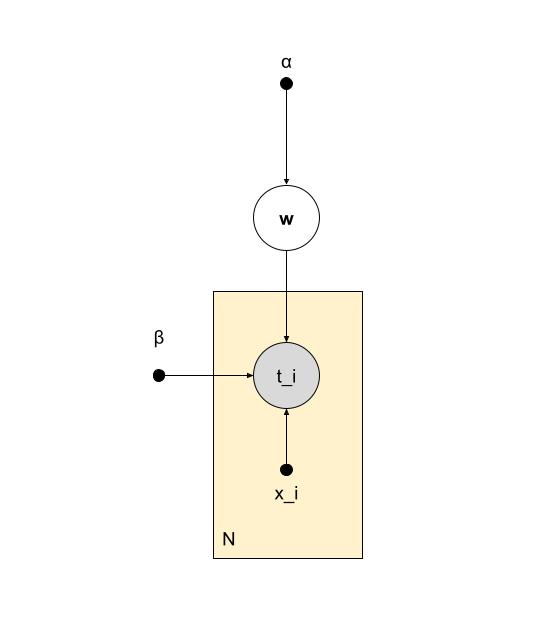
$$ \begin{align*} y_n & \in \mathbb{R} \qquad (output) \\ x_n & \in \mathbb{R}^M \quad e.g. (1,x,x^2,x^3) \qquad (input) \\ \textbf{w} & \in \mathbb{R}^M \qquad (parameter) \\ \epsilon_n & \in \mathbb{R} \qquad (noise) \end{align*} $$
$$ y_n = \textbf{w}^T\textbf{x}_n + \epsilon_n \\ \epsilon_n \sim Norm(\epsilon_n|0,\lambda^{-1}) $$
$$ p(\textbf{t},\textbf{w}|\textbf{x},\alpha^{-1},\beta^{-1}) = p(\textbf{w}|\alpha^{-1})\prod^{\textit{N}}_{n=1}p(\textit{t}_n|\textbf{w},\textit{x}_n,\beta^{-1}) $$
$$ = Norm(\textbf{w}|0,\alpha^{-1})\prod^{\textit{N}}_{n=1}Norm(t_n|\textbf{w}^T\phi(\textit{x}_n),\beta^{-1}) $$
scipy for modeling
M = 3
def phi_basis_fn(x):
result = []
result.append(1.0)
for i in range(M-1):
result.append(x ** (i + 1))
return jnp.array(result)
def model(xx):
# 正定値⾏列なので、独立
mu_w = 0.2
alpha = 0.8
w = stats.norm.rvs(loc=mu_w, scale=1./alpha, size=M)
beta = 0.8
yys, mus = [], []
for x in xx:
mu = jnp.dot(w, phi_basis_fn(x))
yys.append(stats.norm.rvs(loc=mu, scale=1./beta, size=1)[0])
mus.append(mu)
return yys, mus
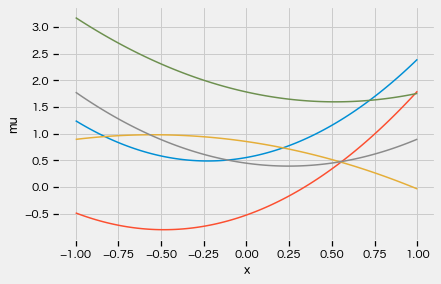
xx = jnp.linspace(-1, 1, 100)
for i in range(5):
_, mu = model(xx)
plt.plot(xx, mu)
plt.xlabel("x")
plt.ylabel("mu")
plt.show()
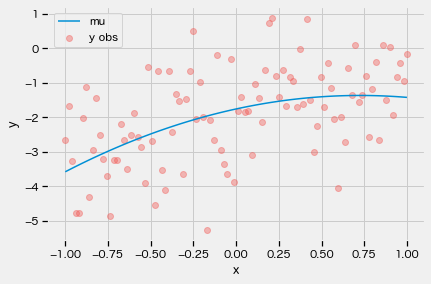
yy, mu = model(xx)
plt.plot(xx, mu, label="mu")
plt.scatter(xx, yy, c=Color[0], alpha=0.4, label="y obs")
plt.xlabel("x")
plt.ylabel("y")
plt.legend()
plt.show()
numpyro
y_obs = jnp.array(yy)
model
def model(y_obs, x_data=None):
mu_w = 0
alpha = 1
ws = numpyro.sample("latent_w", dist.Normal(loc=mu_w, scale=1./alpha), sample_shape=(M,))
N = 1000 if x_data == None else x_data.shape[0]
beta = 4
yys = []
with numpyro.plate("palte", N):
mu = 0
for i in range(M):
mu += ws[i] * (x_data ** i)
y_sample = numpyro.sample("y", dist.Normal(loc=mu, scale=1./beta), sample_shape=(1,), obs=y_obs)
yys.append(y_sample)
return yys
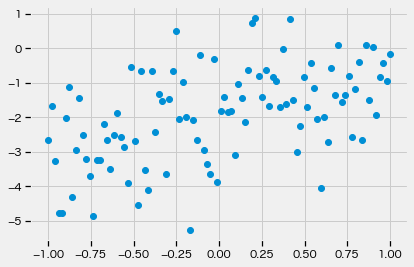
prior_model_trace = handlers.trace(handlers.seed(model, key))
prior_model_exec = prior_model_trace.get_trace(y_obs=y_obs, x_data=xx)
y = prior_model_exec["y"]["value"]
plt.scatter(xx, y)
plt.show()
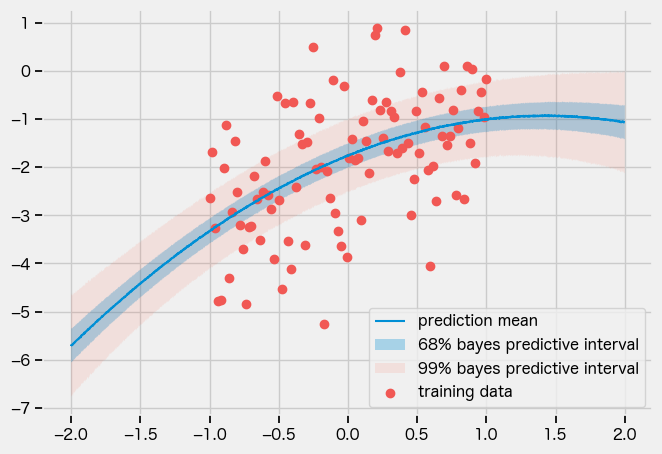
MCMC
kernel = NUTS(model)
mcmc = MCMC(kernel, num_warmup=1000, num_samples=2000)
mcmc.run(key, y_obs=y_obs, x_data=xx)
mcmc.print_summary()
samples = mcmc.get_samples()
predictive = Predictive(model, samples)
idx_pts = jnp.linspace(-2, 2, 2000)
key, subkey = random.split(key)
pred_samples = predictive(subkey, None, idx_pts)["y"]
mean = pred_samples.mean(0)
std = pred_samples.std(0)
lower1 = mean - std
upper1 = mean + std
lower3 = mean - 3*std
upper3 = mean + 3*std
plt.figure(figsize=(7, 5), dpi=100)
plt.plot(idx_pts, mean.squeeze())
plt.fill_between(idx_pts, lower1.squeeze(), upper1.squeeze(), alpha=0.3)
plt.fill_between(idx_pts, lower3.squeeze(), upper3.squeeze(), alpha=0.1)
plt.scatter(xx, y_obs, color=Color[0])
plt.legend(["prediction mean",
"68% bayes predictive interval",
"99% bayes predictive interval",
"training data"])
plt.show()
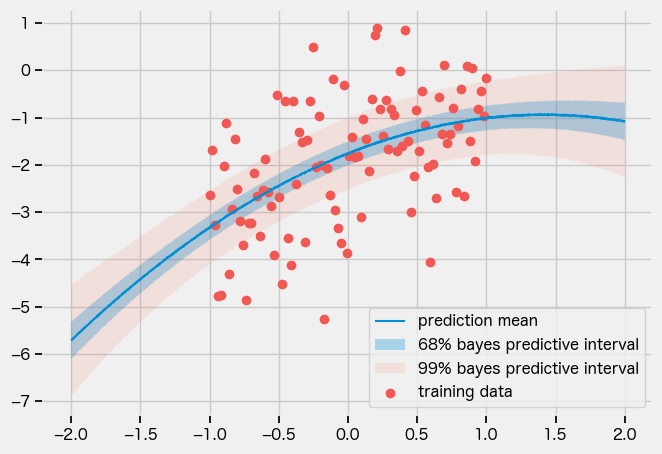
SVI
MAP
guide = numpyro.infer.autoguide.AutoDelta(model)
optimizer = numpyro.optim.Adam(step_size=0.0005)
svi = SVI(model, guide, optimizer, loss=Trace_ELBO())
svi_result = svi.run(key, 5000, y_obs=y_obs, x_data=xx)
params = svi_result.params
pp.pprint(params)
idx_pts = jnp.linspace(-2, 2, 2000)
predictive = Predictive(model=model, guide=guide, params=params, num_samples=idx_pts.shape[0])
key, subkey = random.split(key)
pred_samples = predictive(subkey, y_obs=None, x_data=idx_pts)["y"]
mean = pred_samples.mean(0)
std = pred_samples.std(0)
lower1 = mean - std
upper1 = mean + std
lower3 = mean - 3*std
upper3 = mean + 3*std
plt.figure(figsize=(7, 5), dpi=100)
plt.plot(idx_pts, mean.squeeze())
plt.fill_between(idx_pts, lower1.squeeze(), upper1.squeeze(), alpha=0.3)
plt.fill_between(idx_pts, lower3.squeeze(), upper3.squeeze(), alpha=0.1)
plt.scatter(xx, y_obs, color=Color[0])
plt.legend(["prediction mean",
"68% bayes predictive interval",
"99% bayes predictive interval",
"training data"])
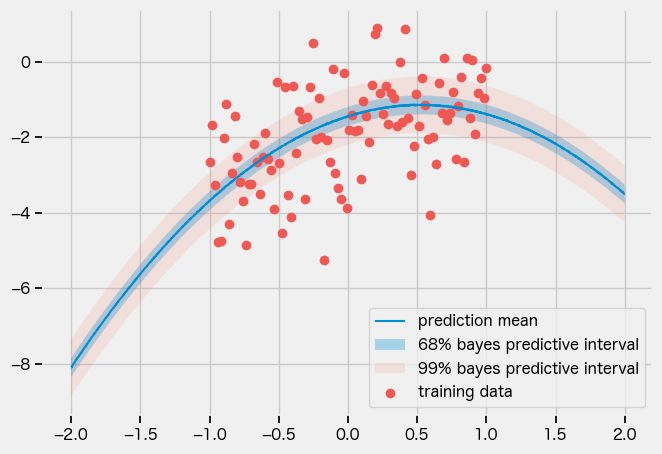
AutoNormal
guide = numpyro.infer.autoguide.AutoNormal(model)
optimizer = numpyro.optim.Adam(step_size=0.0005)
svi = SVI(model, guide, optimizer, loss=Trace_ELBO())
svi_result = svi.run(key, 15000, y_obs=y_obs, x_data=xx)
params = svi_result.params
pp.pprint(params)
idx_pts = jnp.linspace(-2, 2, 2000)
predictive = Predictive(model=model, guide=guide, params=params, num_samples=idx_pts.shape[0])
key, subkey = random.split(key)
pred_samples = predictive(subkey, y_obs=None, x_data=idx_pts)["y"]
mean = pred_samples.mean(0)
std = pred_samples.std(0)
lower1 = mean - std
upper1 = mean + std
lower3 = mean - 3*std
upper3 = mean + 3*std
plt.figure(figsize=(7, 5), dpi=100)
plt.plot(idx_pts, mean.squeeze())
plt.fill_between(idx_pts, lower1.squeeze(), upper1.squeeze(), alpha=0.3)
plt.fill_between(idx_pts, lower3.squeeze(), upper3.squeeze(), alpha=0.1)
plt.scatter(xx, y_obs, color=Color[0])
plt.legend(["prediction mean",
"68% bayes predictive interval",
"99% bayes predictive interval",
"training data"])
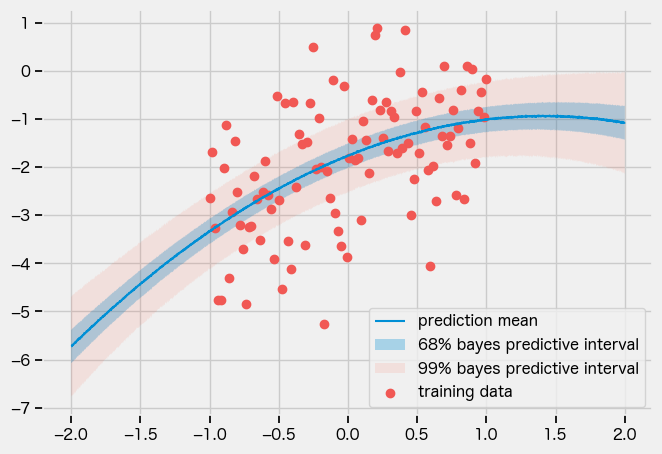
AutoDiagonalNormal
guide = numpyro.infer.autoguide.AutoDiagonalNormal(model)
optimizer = numpyro.optim.Adam(step_size=0.0005)
svi = SVI(model, guide, optimizer, loss=Trace_ELBO())
svi_result = svi.run(key, 15000, y_obs=y_obs, x_data=xx)
params = svi_result.params
pp.pprint(params)
idx_pts = jnp.linspace(-2, 2, 2000)
predictive = Predictive(model=model, guide=guide, params=params, num_samples=idx_pts.shape[0])
key, subkey = random.split(key)
pred_samples = predictive(subkey, y_obs=None, x_data=idx_pts)["y"]
mean = pred_samples.mean(0)
std = pred_samples.std(0)
lower1 = mean - std
upper1 = mean + std
lower3 = mean - 3*std
upper3 = mean + 3*std
plt.figure(figsize=(7, 5), dpi=100)
plt.plot(idx_pts, mean.squeeze())
plt.fill_between(idx_pts, lower1.squeeze(), upper1.squeeze(), alpha=0.3)
plt.fill_between(idx_pts, lower3.squeeze(), upper3.squeeze(), alpha=0.1)
plt.scatter(xx, y_obs, color=Color[0])
plt.legend(["prediction mean",
"68% bayes predictive interval",
"99% bayes predictive interval",
"training data"])
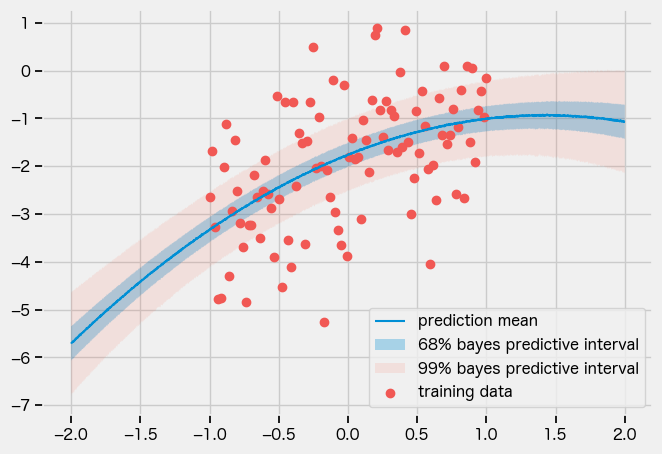
AutoLowRankMultivariateNormal
guide = numpyro.infer.autoguide.AutoLowRankMultivariateNormal(model)
optimizer = numpyro.optim.Adam(step_size=0.0005)
svi = SVI(model, guide, optimizer, loss=Trace_ELBO())
svi_result = svi.run(key, 15000, y_obs=y_obs, x_data=xx)
params = svi_result.params
pp.pprint(params)
idx_pts = jnp.linspace(-2, 2, 2000)
predictive = Predictive(model=model, guide=guide, params=params, num_samples=idx_pts.shape[0])
key, subkey = random.split(key)
pred_samples = predictive(subkey, y_obs=None, x_data=idx_pts)["y"]
mean = pred_samples.mean(0)
std = pred_samples.std(0)
lower1 = mean - std
upper1 = mean + std
lower3 = mean - 3*std
upper3 = mean + 3*std
plt.figure(figsize=(7, 5), dpi=100)
plt.plot(idx_pts, mean.squeeze())
plt.fill_between(idx_pts, lower1.squeeze(), upper1.squeeze(), alpha=0.3)
plt.fill_between(idx_pts, lower3.squeeze(), upper3.squeeze(), alpha=0.1)
plt.scatter(xx, y_obs, color=Color[0])
plt.legend(["prediction mean",
"68% bayes predictive interval",
"99% bayes predictive interval",
"training data"])
full guide
def guide(y_obs, x_data=None):
mu_w_q = numpyro.param("mu_w_q", 0.)
alpha_q = numpyro.param("alpha_q", 1., constraint=constraints.positive)
numpyro.sample("latent_w", dist.Normal(loc=mu_w_q, scale=1./alpha_q), sample_shape=(M,))
optimizer = numpyro.optim.Adam(step_size=0.0005)
svi = SVI(model, guide, optimizer, loss=Trace_ELBO())
svi_result = svi.run(key, 20000, y_obs=y_obs, x_data=xx)
params = svi_result.params
pp.pprint(params)
idx_pts = jnp.linspace(-2, 2, 2000)
predictive = Predictive(model=model, guide=guide, params=params, num_samples=idx_pts.shape[0])
key, subkey = random.split(key)
pred_samples = predictive(subkey, y_obs=None, x_data=idx_pts)["y"]
mean = pred_samples.mean(0)
std = pred_samples.std(0)
lower1 = mean - std
upper1 = mean + std
lower3 = mean - 3*std
upper3 = mean + 3*std
plt.figure(figsize=(7, 5), dpi=100)
plt.plot(idx_pts, mean.squeeze())
plt.fill_between(idx_pts, lower1.squeeze(), upper1.squeeze(), alpha=0.3)
plt.fill_between(idx_pts, lower3.squeeze(), upper3.squeeze(), alpha=0.1)
plt.scatter(xx, y_obs, color=Color[0])
plt.legend(["prediction mean",
"68% bayes predictive interval",
"99% bayes predictive interval",
"training data"])
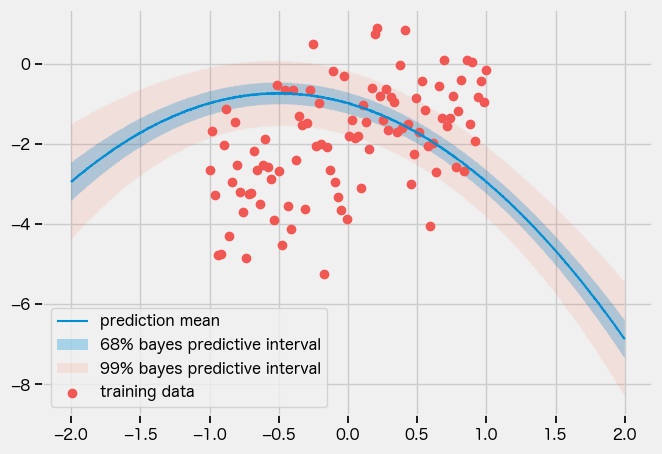
結果
自前のガイドは明らかにおかしいですね。。。なんでかわからん。